For the impatient, you can play with Wooey here (note: you may encounter a server error on the first or second attempt to connect, as the app is hosted on a free plans):
https://wooey.herokuapp.com
And to see the code:
https://www.github.com/wooey/wooey
Wooey, a web UI for your scripts
So, what does Wooey do? Simply put, Wooey takes your scripts:
And converts it to this:
Wooey achieves this by parsing scripts created with command line interface modules, such as argparse. This is achieved through a separate project within the Wooey domain, clinto. Though argparse is really the only command line interface really supported at the moment, we are going to expand clinto to support parsers such as click in the very near future (likely within the next 2 months). Importantly, this is painless for the programmer. A major feature of Wooey is there is no markup language to learn, and users can update scripts through the Django admin in addition to the command line. Scripts are versioned, so jobs and results are reproducible, a much needed feature for any data analysis pipeline.
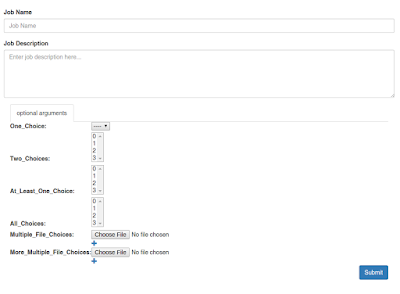
Job Submission with Wooey
As shown above, Wooey provides a simple interface for using a script, and tracks the status of a script's execution and its output:
From here, there are several useful features for any data analysis pipeline:
- Outputs from a script and inspected for common file formats such as tabular data or images, and shown if found.
- Jobs may be shared with users via a unique public url (uuid based, so these are fairly safe against people guessing your job's results) or downloadable as a zip or tar archive
- Jobs may be cloned, which will bring a user to the script submission page with the cloned job's parameters automatically filled in -- thus saving time for running complicated scripts by prepopulating settings.
- Jobs may be rerun or resubmitted to handle cases where a worker node crashed, or other reasons things might just break.
Additionally, for users who create an account, there are more features that allow further customization and ease of use:
- The output of a job can be added to a 'scrapbook', for easy storage of useful output in a centralized place:
- The ability to 'favorite' a script to create your own custom list of commonly utilized tasks.
Other Wooey Features
- For script finding, a user may search for a script:
- All jobs submitted may be viewed in a Datatable, allowing the user to easily find a previous submission:
And probably a few more I missed.
Wooey's Arcitecture
Wooey can be run in several configurations. It can be deployed on a local server with all assets being served by a single machine, but it can also be used on distributed systems with unique configurations such as an ephemeral file system. Thus, Wooey is very scalable for any data analysis needs. To assist in this, we have created several guides for setting up wooey on Heroku, Openshift, and moving various components of Wooey to Amazon (and we are open to pull requests for guides on setting up Wooey on any platform we missed!).
Future plans
There are a ton of plans for Wooey, many which can be seen here. Some notable ones include the ability to load all scripts from a url or a package on pypi, a significantly upgraded user dashboard, more real-time capabilities, and the ability to combine scripts and jobs into a complex workflow.
Contributions
Contributions, suggestions, bug reports, etc. are all welcome on our github repository.